Code outputs¶
Stellar evolution and oscillation programs provide their output
quantities (e.g. luminosity as a function of age or tables of mode
frequencies) in a number of formats. tomso provides functions to make
it easier to work with this data.
Most of the loading functions will interpret filenames starting with
http as URLs and filenames ending with .gz as gzipped and open
them accordingly.
MESA and GYRE¶
MESA’s histories and profiles and GYRE’s summaries and mode outputs
mostly follow a simple text format that can quite easily be read in by
NumPy’s genfromtxt function. Each has a set of scalar quantities
that are fixed for a given output file (e.g. the effective temperature
of a MESA profile or the frequency of a GYRE mode) and a set of
columns for quantities that vary, either over a star’s evolution, as a
function of position in the star, or, for tables of mode frequencies,
as a function of the radial order and angular degree.
tomso’s functions for reading these files return objects that can
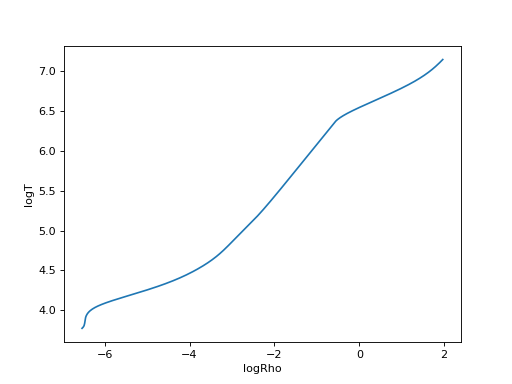
be accessed by keys. e.g. to plot logRho against logT,
import matplotlib.pyplot as pl
from tomso.mesa import load_profile
profile = load_profile('../tests/data/mesa.profile')
pl.plot(profile['logRho'], profile['logT'])
pl.xlabel('logRho')
pl.ylabel('logT')
Note that brackets are stripped so in GYRE files, things like
Re(freq) become Refreq.
The objects for MESA output files also try to make up for logarithmic
keys being absent even if the non-logarithmic form is present (and
vice versa). e.g. you could also access T with profile['T']
even if T is absent, as long as logT (or log_T) is.
Conversely, you can access logT if T is present but logT
isn’t. e.g.:
pl.loglog(profile['Rho'], profile['T'])
will produce the same plot as the previous example even if Rho and
T aren’t columns in the profile data.
ADIPLS¶
ADIPLS produces most of its output in Fortran binary formats and
provides tools to convert these to plain text. tomso provides
functions to read the binary files directly into Python. The binary
output files for the mode frequencies, eigenfunctions and rotation
kernels all have the same basic data for the mode frequencies, which
is stored in ADIPLS as the arrays cs. You can retrieve the full
list of entries from the dtype defined by
tomso.adipls.cs_dtypes. e.g.:
from tomso import adipls
print([name for (name, kind) in adipls.cs_dtypes])
Many of the quantities in the cs arrays are then made available
through relevant properties.
STARS¶
The Cambridge stellar evolution code—STARS—provides two main sets
of plain text output: plot and out. tomso provides associated
routines.
The plot files contains (a lot of!) fixed-width columns of data in
plain-text format. The columns can be accessed using the names defined
in stars.plot_dtypes. e.g.:
from tomso import stars
print([name for (name, kind) in stars.plot_dtypes])
stars.load_plot currently just returns a NumPy record array and
doesn’t (yet) do anything clever with transforming (non-)logarithmic
versions of the columns.
The out files contain a mixture of information, starting with a
copy of the original fixed-format input control file. There are then
regular “summaries”: tables of some stellar properties and, at some
user-defined interval, “profiles” that give the interior properties of
the star. stars.load_out reads a files and returns a
(summaries, profiles) tuple, where each component contains (most
of) the data from the file. The profiles object’s first index is
the profile number; the second index is the row number within that
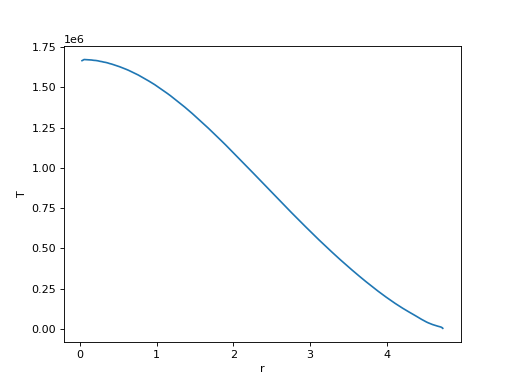
model. So you can plot r against T of the last model
with something like
import matplotlib.pyplot as pl
from tomso.stars import load_out
summaries, profiles = load_out('../tests/data/stars.out')
pl.plot(profiles[-1]['r'], profiles[-1]['T'])
pl.xlabel('r')
pl.ylabel('T')
The profiles’ columns are defined by the user when they run STARS, so
tomso infers their content from the output. You can see what a
profile contains with print(profiles.dtype.names).